Genetically Immortalized Cells: Unlocking the Potential for Scalable Cultured Meat Production
[fusion_builder_container type=”flex” hundred_percent=”no” equal_height_columns=”no” hide_on_mobile=”small-visibility,medium-visibility,large-visibility” background_position=”center center” background_repeat=”no-repeat” fade=”no” background_parallax=”none” parallax_speed=”0.3″ video_aspect_ratio=”16:9″ video_loop=”yes” video_mute=”yes” border_style=”solid”][fusion_builder_row][fusion_builder_column type=”1_1″ layout=”1_1″ background_position=”left top” border_style=”solid” border_position=”all” spacing=”yes” background_repeat=”no-repeat” margin_top=”0px” margin_bottom=”0px” animation_speed=”0.3″ animation_direction=”left” hide_on_mobile=”small-visibility,medium-visibility,large-visibility” center_content=”no” last=”true” hover_type=”none” first=”true” min_height=”” link=”” background_blend_mode=”overlay”][fusion_text columns=”” column_min_width=”” column_spacing=”” rule_style=”default” rule_size=”” rule_color=”” hue=”” saturation=”” lightness=”” alpha=”” content_alignment_medium=”” content_alignment_small=”” content_alignment=”” hide_on_mobile=”small-visibility,medium-visibility,large-visibility” sticky_display=”normal,sticky” class=”” id=”” margin_top=”” margin_right=”” margin_bottom=”” margin_left=”” fusion_font_family_text_font=”” fusion_font_variant_text_font=”” font_size=”” line_height=”” letter_spacing=”” text_transform=”none” text_color=”” animation_type=”” animation_direction=”left” animation_speed=”0.3″ animation_offset=””]
Genetically Immortalized Cells: Unlocking the Potential for Scalable Cultured Meat Production
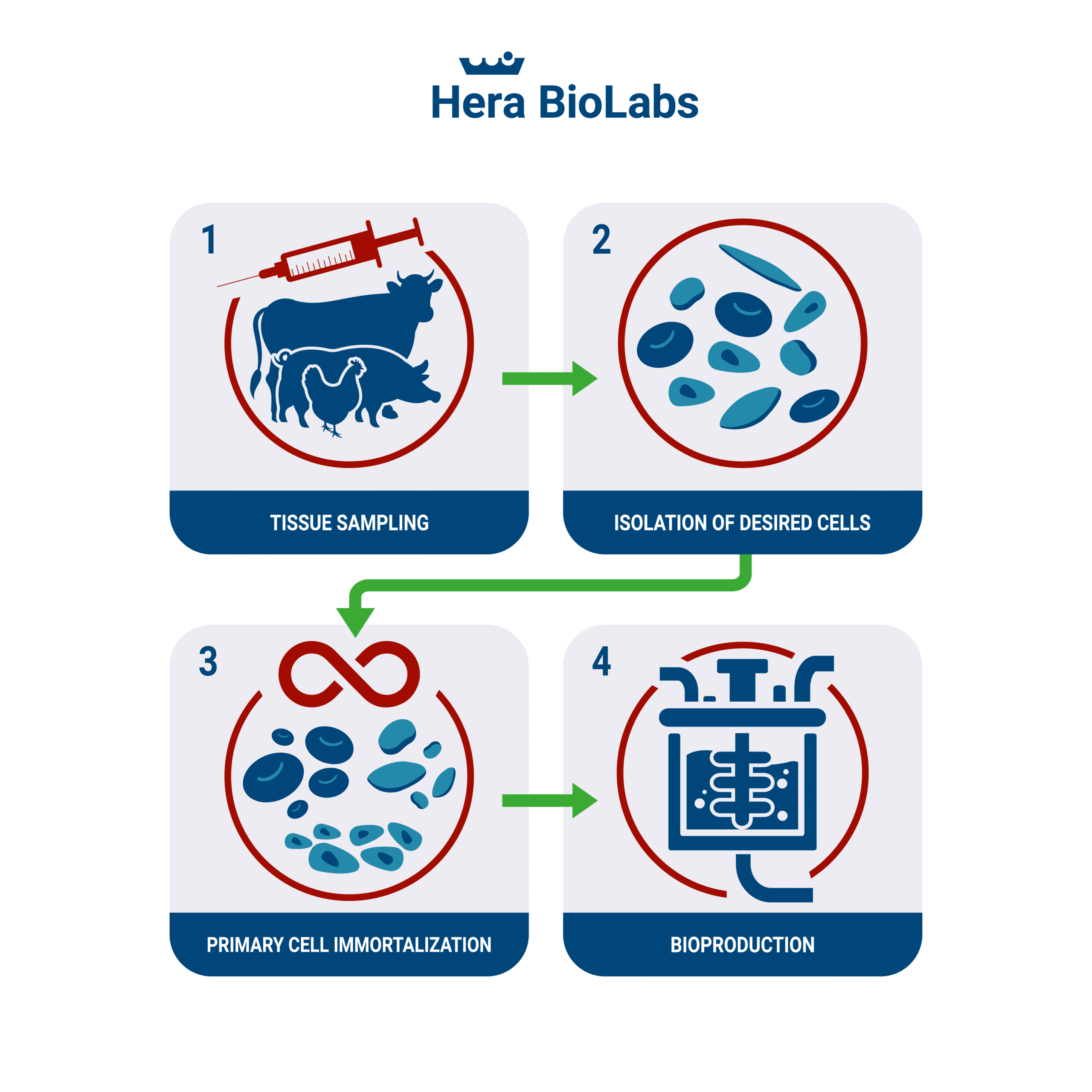
Artificial or cultured meat has been a topic of intense interest for the food industry, with the potential to offer a sustainable and ethically sound alternative to traditional animal farming. Now, a new advancement in this sector is setting the stage for potential scalability: the immortalization of cells for meat production. A recent paper titled “Immortalized Bovine Satellite Cells for Cultured Meat Applications” offers some important insights into how this technology could transform the way we produce meat.
Cultured Meat and Immortalization: A Case Study
Cultured meat, also known as lab-grown meat, is developed by culturing animal cells in a controlled environment. Despite the promise of this technology, one of the major challenges is obtaining a consistent, large-scale source of animal cells that can grow indefinitely, overcoming the Hayflick limit – the natural limit to cellular division. To address this challenge, the paper proposes the idea of genetically immortalized cells. By introducing specific genes that extend a cell’s lifespan, a theoretically infinite supply of cells could be produced. This concept offers significant potential benefits, including rapid growth, bypassing cellular senescence (aging), and a consistent starting cell population for production.
In the study, researchers developed genetically immortalized bovine satellite cells (iBSCs) via constitutive expression of two key components: bovine Telomerase reverse transcriptase (TERT) and Cyclin-dependent kinase 4 (CDK4). Telomerase, an enzyme that lengthens telomeres—the protective ends of chromosomes—allows the cells to divide without experiencing age-related decline. Meanwhile, CDK4, a protein kinase, is crucial for cell cycle progression.
The result was impressive: the iBSCs achieved over 120 doublings at the time of publication and maintained their capacity for myogenic differentiation—meaning they could still differentiate into muscle cells, essential for creating the meat texture. The study’s results thus demonstrate the immense potential of immortalized cells for cultured meat production.
Implications for the Field
The creation of genetically immortalized bovine satellite cells presents an invaluable tool for cultured meat research and development. By providing a reliable and rapidly growing cell population, the technology could enable the mass production of cultured meat, addressing the critical scalability challenge. As biotech companies continue to innovate, the integration of immortalized cells into cultured meat production may not be far from realization. As we continue to refine our methods and improve our understanding of cell biology and genetic engineering, the dream of sustainable, ethical, and scalable meat production comes closer to reality.
PiggyBac for Immortalization
The piggyBac transposon technology is a highly effective approach for cell immortalization, particularly valued for its, efficiency and stability. As a non-viral technology, it also bears significant implications for regulatory approval, offering potential advantages in navigating complex processes like FDA approval.
Here are some key reasons for choosing piggyBac transposon technology in the process of immortalizing cells:
- Non-Viral Delivery System: PiggyBac transposon is a non-viral method of DNA insertion, which presents a notable advantage when considering regulatory aspects. Viral gene delivery methods can sometimes raise safety concerns due to their potential for mutagenesis and immunogenicity. On the contrary, piggyBac, as a non-viral technology, is less likely to trigger these concerns, which can streamline the process of securing regulatory approval, such as from the FDA.
- High Carrying Capacity: PiggyBac transposons can carry large genetic elements, enabling the insertion of multiple genes or extensive genetic components. This is crucial when multiple genes are required for cell immortalization. The piggyBac cargo capacity extends beyond 25 kb in size.
- Stable Gene Expression: Genes introduced using piggyBac transposition maintain stable expression in the cell. This stability is vital for cell immortalization, where continuous expression of specific genes is necessary.
The piggyBac transposon technology offers an easy non-viral solution for gene transfer and cell immortalization. Contact Hera if you are interested in using piggyBac for cell line engineering or immortalization products.
[/fusion_text][/fusion_builder_column][/fusion_builder_row][/fusion_builder_container]