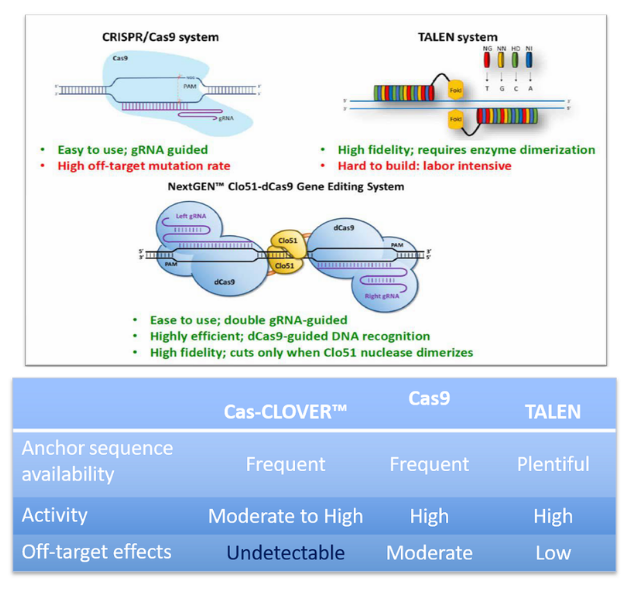
CRISPR/Cas9 has set standard for efficient gene editing in non-commercial settings. However, access is an issue for many users due to a lack of commercial freedom to operate in CRISPR’s licensing. In addition, CRISPR/Cas9’s reliance on a single guide RNA has been shown to produce “off-target” mutations – genomic cuts in unwanted sites.
On the other hand, the high-precision dimeric Cas-CLOVER gene editing technology is supported by simple patents and has demonstrated zero detected off-target activity. This “proven alternative to CRISPR/Cas9” was recently validated in CHO cells, and maintains the high efficiency and ease of use advantages of CRISPR/Cas9.
Hera BioLabs offers commercial and academic licenses as well as proof of concept services for this technology in drug discovery & early development. Schedule a call to learn more.
Why Cas-CLOVER?
- It is functionally similar to other CRISPR/Cas9 technologies but uses a different nuclease protein called Clo51, which is covered under a set of patents distinct from other CRISPR/Cas9 technologies.
- It achieves greater specificity through the utilization of two guide RNAs as well as a nuclease activity that requires the dimerization of subunits associated with each guide RNA.
- It is an exclusive biotechnology that is only sub-licensable by Hera for drug discovery and early development.
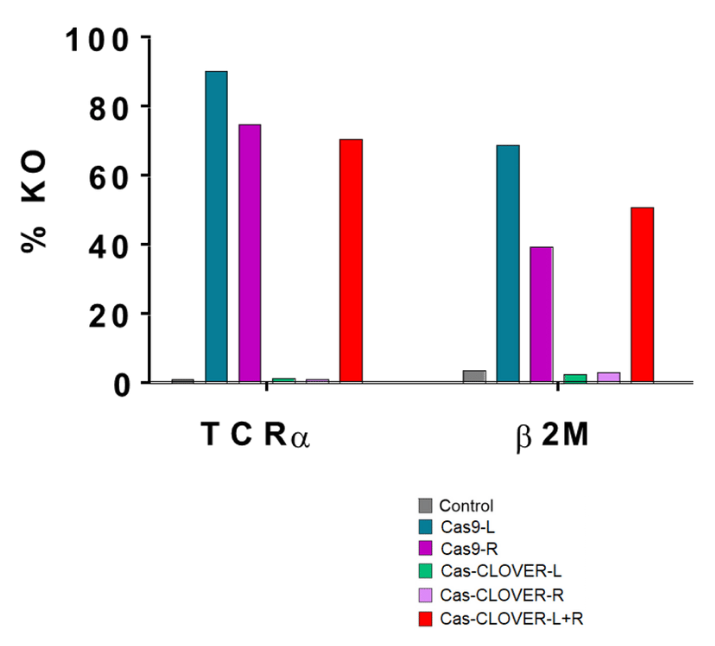
Cas-CLOVER Activity is Comparable to Cas9
Side-by-side comparison of CRISPR/Cas9 or Cas-CLOVER disruption of surface TCR expression in resting human T cells as assessed by FACS. Single or multiplexed (left (L) + right (R)) TCRα-specific gRNAs were tested.
In contrast to CRISPR/Cas9, Cas-CLOVER requires both L and R gRNAs for on-site targeting. Additional targets were tested (β2M) and data is summarized by this bar graph
Cas-CLOVER demonstrated comparable efficiency to CRISPR/Cas9 in disrupting the target gene.
Cas-CLOVER’s requirement of two guide RNAs with activity dependent upon relatively strict spacer lengths results in a highly specific genome editing tool.
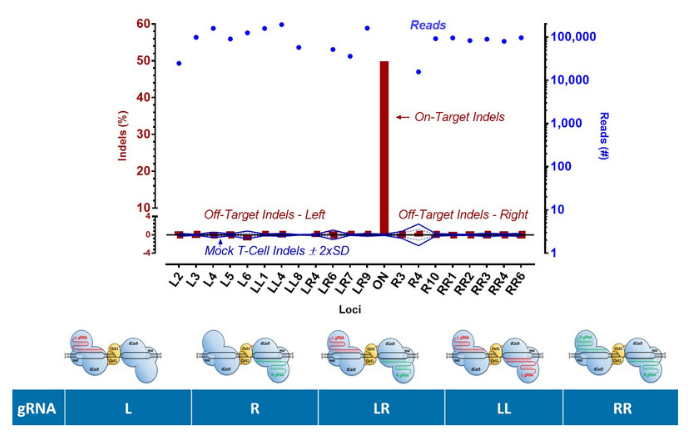
Cas-CLOVER: No Off Targets Detected By NGS
PD1 target knockouts by Cas-CLOVER system were conducted in resting T cells. Resting T cells without Cas-CLOVER (3 replicates) were used as a control. Genomic DNA was collected and the top predicted off-target sites were selected for target amplicon amplification and sequencing by NGS.
No off-target mutations above background were observed by NGS sequencing, with most of the sites at 30,000-100,000X coverage. Possible combinations of gRNAs with Cas-CLOVER as shown.